PDX model details
| PDX ID |
373M |
| Host Strain(and Source) |
NSG
Source: Monash University |
| Host Strain Immune system Humanized |
NO |
| Host Type |
Testosterone supplemented |
| Graft Site |
Subcutaneous |
Current Generation
(* indicates number of generations grown in Castrate host) |
13 |
| Average PDX Generation Time (days +/- SEM) |
78 ± 7 |
| Tumour preparation |
Tumor solid |
| Tumour Characterization Technology |
Histology, IHC, STR profiling, targeted DNA sequencing, RNAseq |
| Tumour confirmed not to be of Mouse/EBV origin |
Yes; negative for CD45 and/or positive for human CK8/18 and/or human mitochondria by IHC |
| Passage QA performed |
Routine QA every 2-3 passages |
| Associated meta data |
| PDX model availability |
Yes (fixed, frozen) |
| Governance restriction for distribution |
Available to investigators with appropriate ethics approvals, approved EOI from MURAL committee, and material transfer agreements |
| Pubmed ID |
34413304 |
| |
|
| Markers |
373M |
| AR |
N |
| PSA |
N |
| PSMA |
N |
| NE |
Y |
| ERG |
N |
This heatmap displays the call type of curated CNVs in the presented samples.
This heatmap displays the call type of CNVs in commonly occurred samples.
This table displays curated CNVs
|
PDX ID
|
Gene Symbol
|
CNV Log2
|
CNV Copy
|
CNV Call
|
Experiments name
|
Platform
|
Reference genome
|
| 373M |
MYC |
0.559469 |
2.94745 |
gain
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
SMAD2 |
0.888683 |
3.70297 |
gain
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
FANCD2 |
-0.746724 |
1.19191 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
RAF1 |
-0.746724 |
1.19191 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
SETD2 |
-0.759832 |
1.18113 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
PIK3R1 |
-1.63343 |
0.644642 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
RB1 |
-1.62166 |
0.649923 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
CDH1 |
-0.837756 |
1.11903 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
ZFHX3 |
-0.837756 |
1.11903 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
FANCA |
-0.837756 |
1.11903 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
| 373M |
TP53 |
-1.19018 |
0.876496 |
loss
|
MURAL Prostate Cancer PDX collection |
Targeted GARVAN Panel |
hg19 |
Clinical Information
| Sample Number |
373M |
| Sample Site |
Liver |
| Sample source |
Biopsy |
| Pathology Tumour Diagnosis |
Mixed Adeno & Neuro |
| Gleason Score |
None |
| Primary Gleason Score |
None |
| Secondary Gleason Score |
None |
| Tertiary Gleason Score |
None |
| ISUP Grade Group |
|
| Tumour Grade |
|
| D'Amico Risk Classification |
|
| Tumour Volume (in cc) |
0.0 |
| Treatment Prior to Specimen Collection |
ADT |
| |
|
Patient Information
| Patient Number |
373 |
| Sex |
Male |
| Diagnosis |
Prostate Cancer |
| PSA at diagnosis (ng/mL) |
NA |
| Consent to share data |
|
| |
|
This heatmap displays the mutations of curated sequence variants.
Download
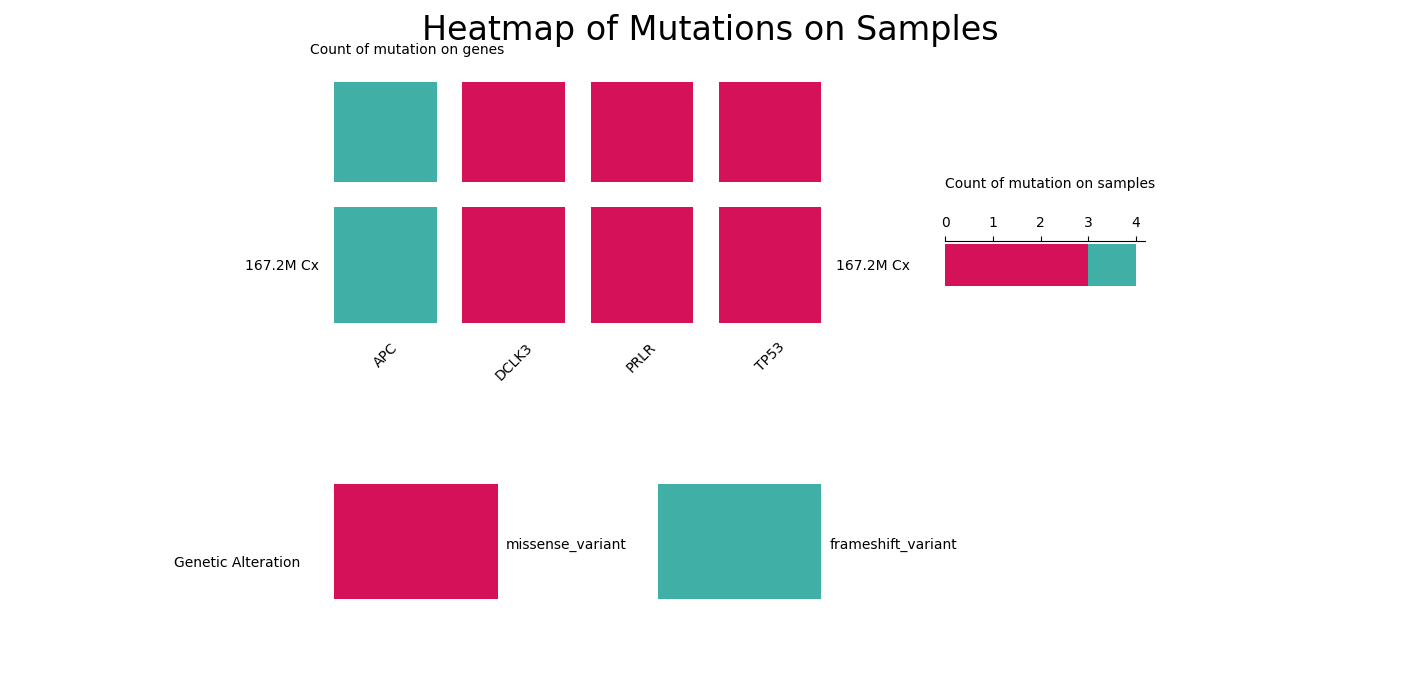
This table displays curated sequence variants
|
PDX ID
|
Gene Symbol
|
Depth
|
ALT_FREQ
|
Consequence
|
Exon
|
GnomAD_AF
|
CADD_PHRED
|
Clinvar_clnsig
|
Platform
|
Experiments Name
|
Reference
|
Library Type
|
Instrument Type
|
| 373M |
RB1 |
19 |
1.0 |
missense_variant,splice_region_variant |
'15/27 |
. |
33 |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
TP53 |
119 |
0.97 |
missense_variant |
'8/11 |
0 |
28.8 |
Conflicting_interpretations_of_pathogenicity
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
MAP3K1 |
62 |
0.71 |
inframe_insertion |
'1/20 |
0 |
. |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
MAP2K2 |
143 |
0.59 |
missense_variant |
'9/11 |
. |
22 |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
ABL1 |
497 |
0.51 |
missense_variant |
'11/11 |
0.00013 |
28.6 |
not_provided
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
ADAMTS7 |
68 |
0.44 |
missense_variant |
'7/24 |
. |
26.3 |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
MYO5A |
254 |
0.35 |
missense_variant |
'15/41 |
6.50E-05 |
24.4 |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
SOS1 |
196 |
0.3 |
missense_variant |
'4/24 |
0.000114 |
32 |
Uncertain_significance
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
XPO1 |
200 |
0.3 |
missense_variant |
'15/25 |
4.47E-06 |
34 |
Likely_pathogenic
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
| 373M |
SMO |
482 |
0.28 |
missense_variant |
'3/12 |
0.000111 |
28.6 |
-
|
Targeted GARVAN Panel |
MURAL Prostate Cancer PDX collection |
hg19 |
paired |
Illumina HiSeq 2500 |
No information in Drug dosing Table

