PDX model details
PDX ID
426M
Host Strain(and Source)
NSG
Host Strain Immune system Humanized
NO
Host Type
Testosterone supplemented
Graft Site
Subcutaneous
Current Generation
6
Average PDX Generation Time (days +/- SEM)
83 ± 5
Tumor preparation
Tumor solid
Tumor Characterization Technology
Histology, IHC, STR profiling, targeted DNA sequencing, RNAseq
Tumor confirmed not to be of Mouse/EBV origin
Yes; negative for CD45 and/or positive for human CK8/18 and/or human mitochondria by IHC
Passage QA performed
Routine QA every 2-3 passages
Associated meta data
PDX model availability
Yes (fixed, frozen)
Governance restriction for distribution
Available to investigators with appropriate ethics approvals, approved EOI from MURAL committee, and material transfer agreements
Pubmed ID
34413304
Markers
426M
AR
N
PSA
N
PSMA
N
NE
Y
ERG
N
This heatmap displays the call type of curated CNVs in the presented samples.
This heatmap displays the call type of CNVs in commonly occurred samples.
This table displays curated CNVs
Clinical Information
Sample Number
426M
Sample Site
Lung
Sample source
Surgery
Pathology Tumor Diagnosis
None
Gleason Score
None
Primary Gleason Score
None
Secondary Gleason Score
None
Tertiary Gleason Score
None
ISUP Grade Group
Tumor Grade
D'Amico Risk Classification
Tumor Volume (in cc)
0.0
Treatment Prior to Specimen Collection
PRRT LuTate and Y-Tate, Octreotide
Patient Information
Patient Number
426/452
Sex
Male
Diagnosis
Prostate Cancer
PSA at diagnosis (ng/mL)
NA
Consent to share data
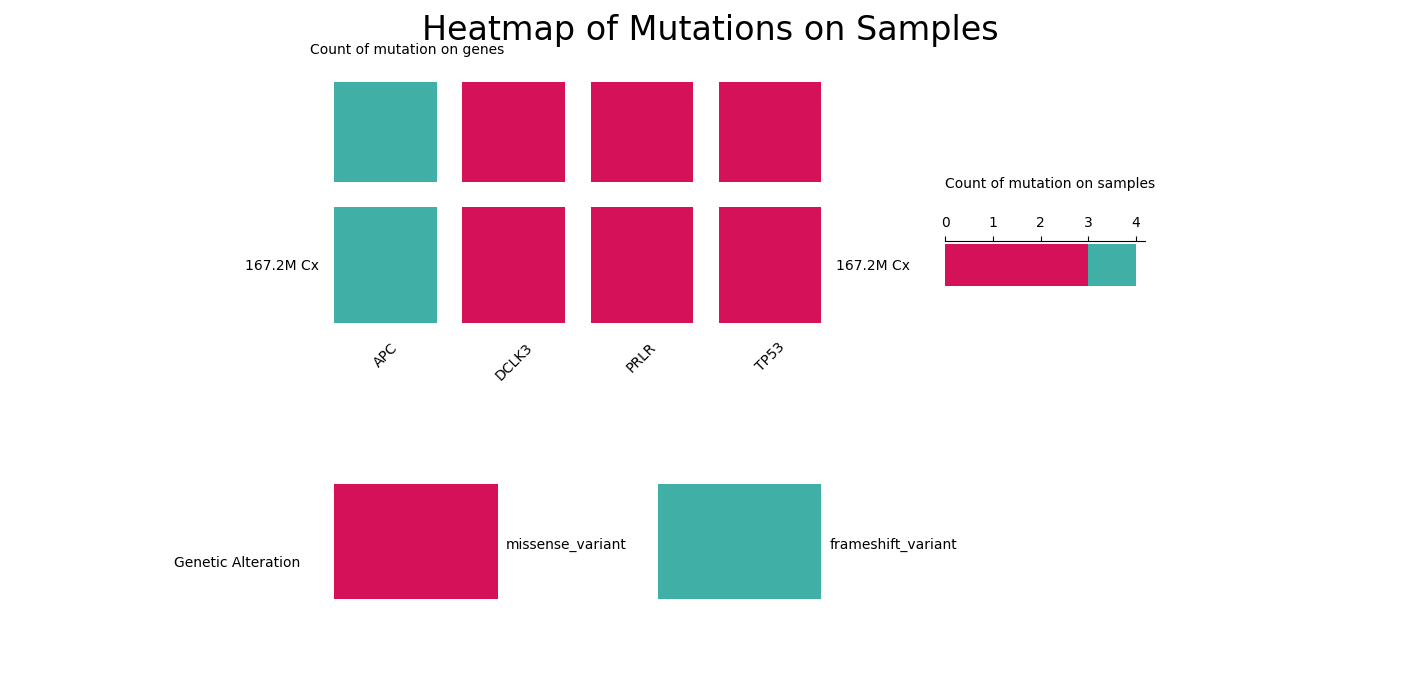
This heatmap displays the mutations of curated sequence variants.
Download
This table displays curated sequence variants
PDX ID
Gene Symbol
Depth
ALT_FREQ
Consequence
Exon
GnomAD_AF
CADD_PHRED
Clinvar_clnsig
Platform
Experiments Name
Reference
Library Type
Instrument Type
426M
DICER1
406
0.68
missense_variant
'25/29
.
27.9
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
FOXA1
380
0.64
missense_variant
'2/2
4.07E-06
25.4
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
BRCA1
341
0.55
missense_variant
'7/24
0.000264
24.7
Benign
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
BRCA1
459
0.51
missense_variant
'10/24
0.0002605
17.67
Benign
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
BRCA1
442
0.48
missense_variant
'10/24
0.0002845
0.005
Benign
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
LATS2
335
0.48
missense_variant
'8/8
4.12E-06
25.3
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
FOXA2
508
0.47
stop_gained
'2/2
.
48
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
FOXA2
508
0.47
stop_gained
'2/2
.
48
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
NCOR1
460
0.46
missense_variant
'25/46
0.0006336
22.7
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
426M
FGFR3
621
0.27
missense_variant
'13/18
.
25.6
-
Targeted Twist Panel
MURAL Prostate Cancer PDX collection
hg19
paired
NextSeq 500
No information in Drug dosing Table
 A
web-based Atlas of Patient-Derived Xenografts
of cancer patients:A resource of MURAL PDX data
A
web-based Atlas of Patient-Derived Xenografts
of cancer patients:A resource of MURAL PDX data